Sponsor:
Advanced Scientific Computing Research (ASCR) under the U.S. Department of Energy Office of Science (Office of Science)
Project Team Members:
Northwestern University
The HDF Group
- Quincey Koziol
- Gerd Herber
Argonne National Laboratory
North Carolina State University
- Nagiza Samatova
- Sriram Lakshminarasimhan
Damsel Usecase - Subfiling
Overview
Subfiling is a mechanism to partition the damsel file into multiple partitioned files (subfiles) internally, making the damsel data appear as a single file to users. Currently user's intent of using subfiling is conveyed through the damsel trait. Once this information is given to the damsel library, all tags defined in the program will be partitioned and stored into subfiles.
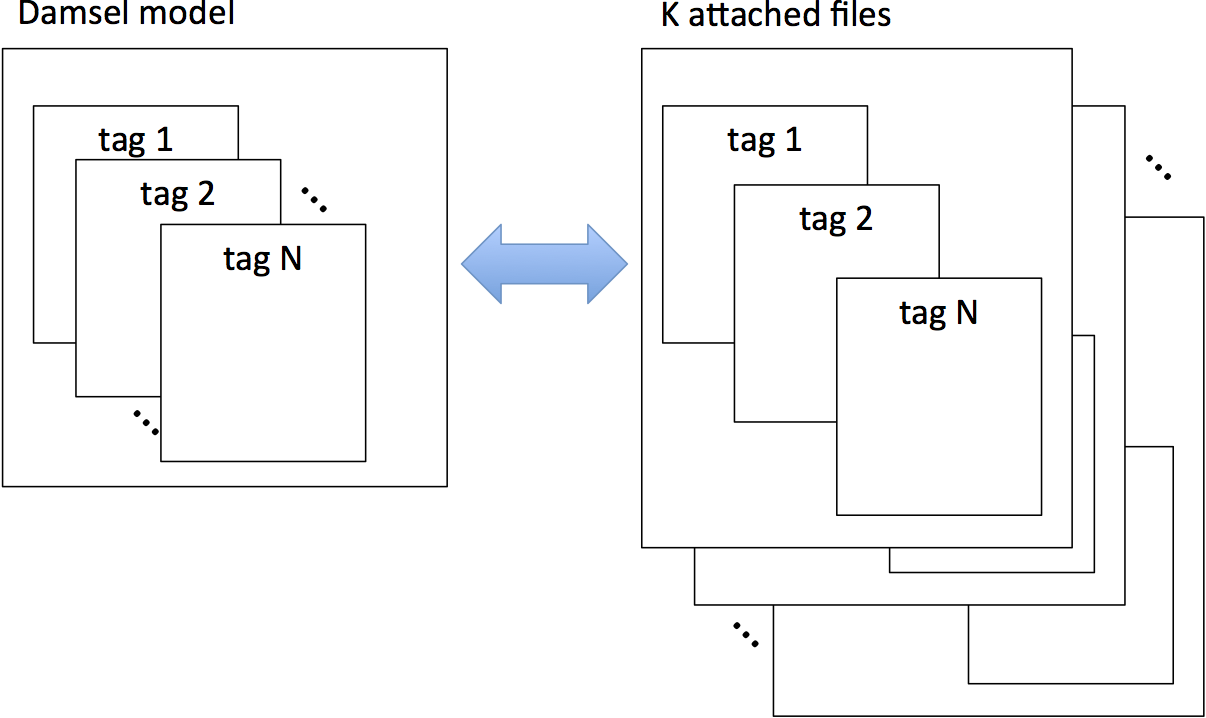
Figure below illustrates a high-level view of the subfiling mechanism. In contrary to a normal case where a damsel model has only one attached damsel file, subfiling creates multiple attached damsel files to a damsel model.
Building and running with subfiling
The subfiling is disabled by default. To enable it, the damsel needs to be configured explicitly using --enable-subfiling.
Once it is enabled, users should convey their intention of using subfiling through the damsel trait:
... damsel_trait_bundle tb; DMSLtrait_create(&tb); DMSLtrait_put(tb, "mode", "WRITE"); DMSLtrait_put(tb, "dmsl_num_subfiles", "2"); DMSLmodel_attach(model, FILENAME, MPI_COMM_WORLD, tb); ...
The example above will create two subfiles attached to user's damsel file. During writes, all subfile related information is stored in the master file, so that reading from subfiles is can be done transparently. In other words, there will be no code change in reading cases.
Note that the programs built with subfiling should be run in parallel, that is running with 2 or more MPI ranks. This applies to both writes and read cases. Note also that users cannot specify the number of subfiles higher than the number of MPI ranks because during subfiing the original communicator is partitioned according to the number of subfiles. If the number of subfiles is greater than the available MPI ranks, the program will create normal file without partitioning.
File layout with/without subfiling
This section describes the file layout with and without subfiling to help understand the mechanism behind subfiling module in damsel. Note that regardless of whether subfiling is enabled or not, all files generated are in the damsel file format. The default damsel file format comprises of two types of datasets: one for storing metadata and the other for storing actual data. If no subfiling is used, all datasets are stored in a single file specified by the user. For example, the .h5 file, nFilewrite.h5, generated by running test-nwrite.c using 2 ranks looks like:
HDF5 "output/nFilewrite.h5" {
GROUP "/" {
DATASET "SolData-200001395" {
DATATYPE H5T_STD_I16LE
DATASPACE SIMPLE { ( 5 ) / ( 5 ) }
DATA {
(0): 0, 11, 22, 33, 44
}
}
DATASET "SolData-300001395" {
DATATYPE H5T_STD_I16LE
DATASPACE SIMPLE { ( 5 ) / ( 5 ) }
DATA {
(0): 1, 12, 23, 34, 45
}
}
In above datasets, SolData-200001395 and SolData-300001395 are the data portion belonging to rank 0 and rank 1, respectively.
If the number of subfiles is set to N, the same program will generate N+1 files. For example, the above test case will generate one master (nFilewrite.h5) and two subfiles (nFilewrite.h5.subfile_0 and nFilewrite.h5.subifle_1). Since the file is partitioned into 2, now the original (or master) file does not contain any datasets but includes the information associated with subfiles. Those information is stored as two metadata, damsel-num-subfiles and damsel-subfile-range. damsel-num-subfiles is a metadata that specify the number of subfiles associated with the damsel file. damsel-subfile-range is on the other hand for specifying the data range stored in each subfile.
Using the example above, the subfiling-related content of nFilewrite.h5 is:
HDF5 "output/nFilewrite.h5" {
GROUP "/" {
DATASET "SolData-200001395-len" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 1 ) / ( 1 ) }
DATA {
(0): 5
}
}
DATASET "SolData-300001395-len" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 1 ) / ( 1 ) }
DATA {
(0): 5
}
}
DATASET "damsel-num-subfiles" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 1 ) / ( 1 ) }
DATA {
(0): 2
}
}
Each subfile contains its dataset as follows:
HDF5 "output/nFilewrite.h5.subfile_0" {
GROUP "/" {
DATASET "SolData-200001395" {
DATATYPE H5T_STD_I16LE
DATASPACE SIMPLE { ( 5 ) / ( 5 ) }
DATA {
(0): 0, 11, 22, 33, 44
}
}
HDF5 "output/nFilewrite.h5.subfile_1" {
GROUP "/" {
DATASET "SolData-300001395" {
DATATYPE H5T_STD_I16LE
DATASPACE SIMPLE { ( 5 ) / ( 5 ) }
DATA {
(0): 1, 12, 23, 34, 45
}
}
test-subfile-parallel.sh is the test script for subfiling. Currently, the test is based on test-nwrite.c, test-parallel-read.c, test-parallel-write.c, test-flash-write-parallel.c and test-nconnectivitywrite.c.

|
EECS Home |
McCormick Home |
Northwestern Home |
Calendar: Plan-It Purple © 2011 Robert R. McCormick School of Engineering and Applied Science, Northwestern University "Tech": 2145 Sheridan Rd, Tech L359, Evanston IL 60208-3118 | Phone: (847) 491-5410 | Fax: (847) 491-4455 "Ford": 2133 Sheridan Rd, Ford Building, Rm 3-320, Evanston, IL 60208 | Fax: (847) 491-5258 Email Director Last Updated: $LastChangedDate: 2016-11-06 13:53:06 -0600 (Sun, 06 Nov 2016) $ |